(labels, /*sx=*/512, /*sy=*/512, /*sz=*/512, source);
```
## Python `pip` Binary Installation
```bash
pip install dijkstra3d
```
## Python `pip` Source Installation
*Requires a C++ compiler.*
```bash
pip install numpy
pip install dijkstra3d
```
## Python Direct Installation
*Requires a C++ compiler.*
```bash
git clone https://github.com/seung-lab/dijkstra3d.git
cd dijkstra3d
virtualenv -p python3 venv
source venv/bin/activate
pip install -r requirements.txt
python setup.py develop
```
## Performance
I ran three algorithms on a field of ones from the bottom left corner to the top right corner of a 512x512x512 int8 image using a 3.7 GHz Intel i7-4920K CPU. Unidirectional search takes about 42 seconds (3.2 MVx/sec) with a maximum memory usage of about 1300 MB. In the unidirectional case, this test forces the algorithm to process nearly all of the volume (dijkstra aborts early when the target is found). In the bidirectional case, the volume is processed in about 11.8 seconds (11.3 MVx/sec) with a peak memory usage of about 2300 MB. The A* version processes the volume in 0.5 seconds (268.4 MVx/sec) with an identical memory profile to unidirectional search. A* works very well in this simple case, but may not be superior in all configurations.
Theoretical unidirectional memory allocation breakdown: 128 MB source image, 512 MB distance field, 512 MB parents field (1152 MB). Theoretical bidirectional memory allocation breakdown: 128 MB source image, 2x 512 distance field, 2x 512 MB parental field (2176 MB).

Fig. 1: A benchmark of dijkstra.dijkstra run on a 5123 voxel field of ones from bottom left source to top right target. (black) unidirectional search (blue) bidirectional search (red) A* search aka compass=True.
```python
import numpy as np
import time
import dijkstra3d
field = np.ones((512,512,512), order='F', dtype=np.int8)
source = (0,0,0)
target = (511,511,511)
path = dijkstra3d.dijkstra(field, source, target) # black line
path = dijkstra3d.dijkstra(field, source, target, bidirectional=True) # blue line
path = dijkstra3d.dijkstra(field, source, target, compass=True) # red line
```
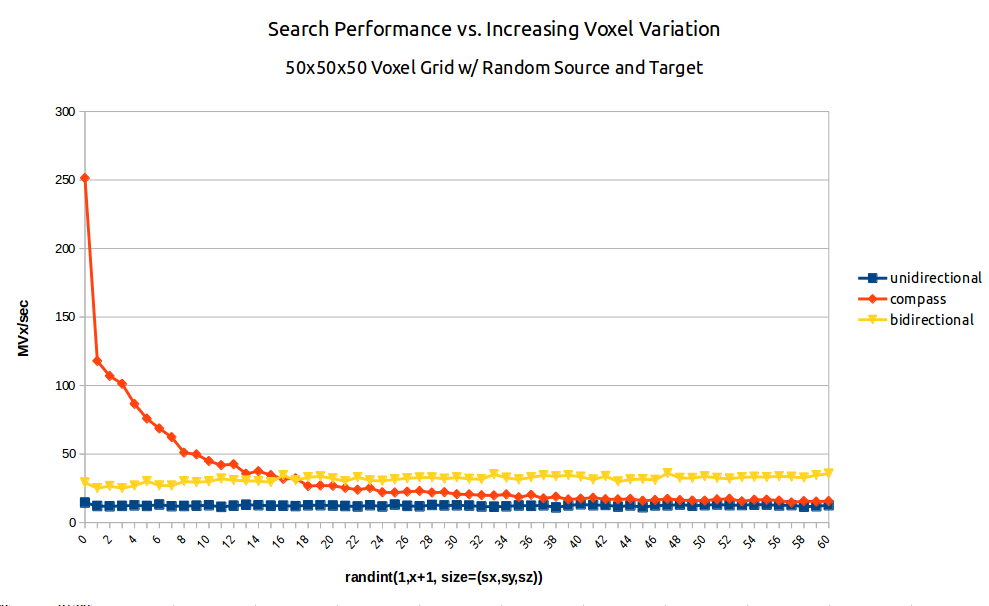
Fig. 2: A benchmark of dijkstra.dijkstra run on a 503 voxel field of random integers of increasing variation from random source to random target. (blue/squares) unidirectional search (yellow/triangles) bidirectional search (red/diamonds) A* search aka compass=True.
```python
import numpy as np
import time
import dijkstra3d
N = 250
sx, sy, sz = 50, 50, 50
def trial(bi, compass):
for n in range(0, 100, 1):
accum = 0
for i in range(N):
if n > 0:
values = np.random.randint(1,n+1, size=(sx,sy,sz))
else:
values = np.ones((sx,sy,sz))
values = np.asfortranarray(values)
start = np.random.randint(0,min(sx,sy,sz), size=(3,))
target = np.random.randint(0,min(sx,sy,sz), size=(3,))
s = time.time()
path_orig = dijkstra3d.dijkstra(values, start, target, bidirectional=bi, compass=compass)
accum += (time.time() - s)
MVx_per_sec = N * sx * sy * sz / accum / 1000000
print(n, ',', '%.3f' % MVx_per_sec)
print("Unidirectional")
trial(False, False)
print("Bidirectional")
trial(True, False)
print("Compass")
trial(False, True)
```
## Voxel Connectivity Graph
You may optionally provide a unsigned 32-bit integer image that specifies the allowed directions of travel per voxel as a directed graph. Each voxel in the graph contains a bitfield of which only the lower 26 bits are used to specify allowed directions. The top 6 bits have no assigned meaning. It is possible to use smaller width bitfields for 2D images (uint8) or for undirected graphs (uint16), but they are not currently supported. Please open an Issue or Pull Request if you need this functionality.
The specification below shows the meaning assigned to each bit. Bit 32 is the MSB, bit 1 is the LSB. Ones are allowed directions and zeros are disallowed directions.
```
32 31 30 29 28 27 26 25 24 23
------ ------ ------ ------ ------ ------ ------ ------ ------ ------
unused unused unused unused unused unused -x-y-z x-y-z -x+y-z +x+y-z
22 21 20 19 18 17 16 15 14 13
------ ------ ------ ------ ------ ------ ------ ------ ------ ------
-x-y+z +x-y+z -x+y+z xyz -y-z y-z -x-z x-z -yz yz
12 11 10 9 8 7 6 5 4 3
------ ------ ------ ------ ------ ------ ------ ------ ------ ------
-xz xz -x-y x-y -xy xy -z +z -y +y
2 1
------ ------
-x +x
```
There is an assistive tool available for producing these graphs from adjacent labels in the [cc3d library](https://github.com/seung-lab/connected-components-3d).
### What is that pairing_heap.hpp?
Early on, I anticipated using decrease key in my heap and implemented a pairing heap, which is supposed to be an improvement on the Fibbonacci heap. However, I ended up not using decrease key, and the STL priority queue ended up being faster. If you need a pairing heap outside of boost, check it out.
## References
1. E. W. Dijkstra. "A Note on Two Problems in Connexion with Graphs" Numerische Mathematik 1. pp. 269-271. (1959)
2. E. W. Dijkstra. "Go To Statement Considered Harmful". Communications of the ACM. Vol. 11, No. 3, pp. 147-148. (1968)
3. Pohl, Ira. "Bi-directional Search", in Meltzer, Bernard; Michie, Donald (eds.), Machine Intelligence, 6, Edinburgh University Press, pp. 127-140. (1971)
